QUAST-LG is an extension of QUAST intended for evaluating large-scale genome assemblies (up to mammalian-size).
Significant speedup of the tool was achieved by both use of new fast aligner (minimap2) and the refactoring of core QUAST alignment analyzing modules.
QUAST-LG also introduced new quality metrics and methods which should be useful when evaluating large eukaryotic assemblies:
- K-mer-based coompleteness and correctness
- Number of full and partial BUSCO genes (especially helpful in the reference-free mode)
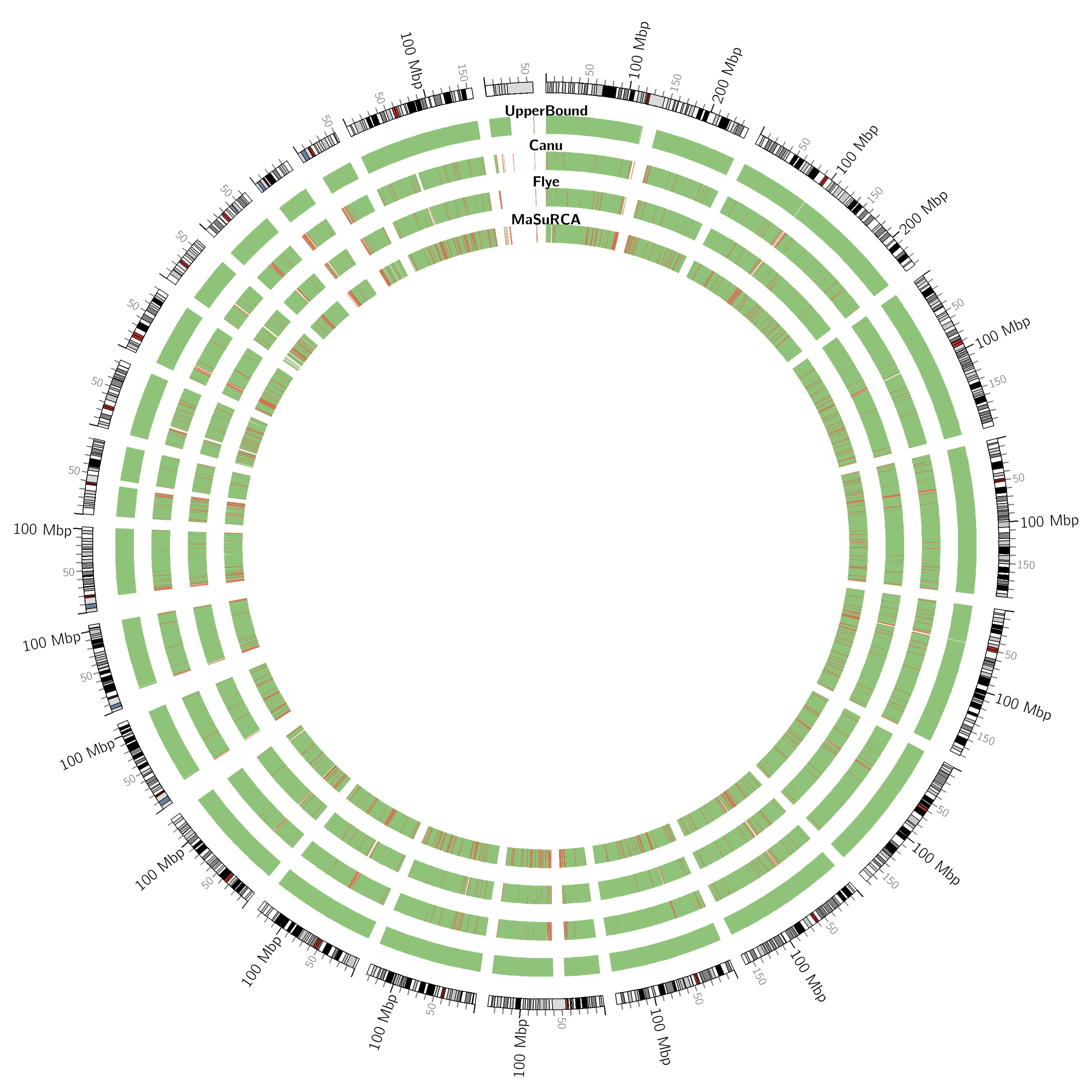
- The concept of upper bound assembly (theoretical limits on the assembly completeness and contiguity for a given genome and set of reads)
QUAST-LG is distributed as a command-line tool within QUAST package since version 5.0.0.
To run QUAST in QUAST-LG mode you should just add --large option to your running command.
Citation:
Versatile genome assembly evaluation with QUAST-LG,
Bioinformatics (2018) 34 (13): i142-i150. doi: 10.1093/bioinformatics/bty266
First published online: June 27, 2018
We benchmarked QUAST-LG on six eukaryotic datasets (yeast [2], worm [1], fruit fly [1], and human [2]) assembled by the leading genome assembly software.
You may find additional information about the results and the QUAST-LG project on the ISMB-2018 poster.
Below is the sample plot generated in QUAST-LG run on a human dataset (using Circos and Icarus software).
Click on it to go to the full report on this dataset.
Please help us make QUAST better by sending your comments, bug reports, and suggestions
to alexey.gurevich@helmholtz-hips.de or
posting them on our GitHub issue tracker
(please check that an issue similar to yours is not solved or explained already!).