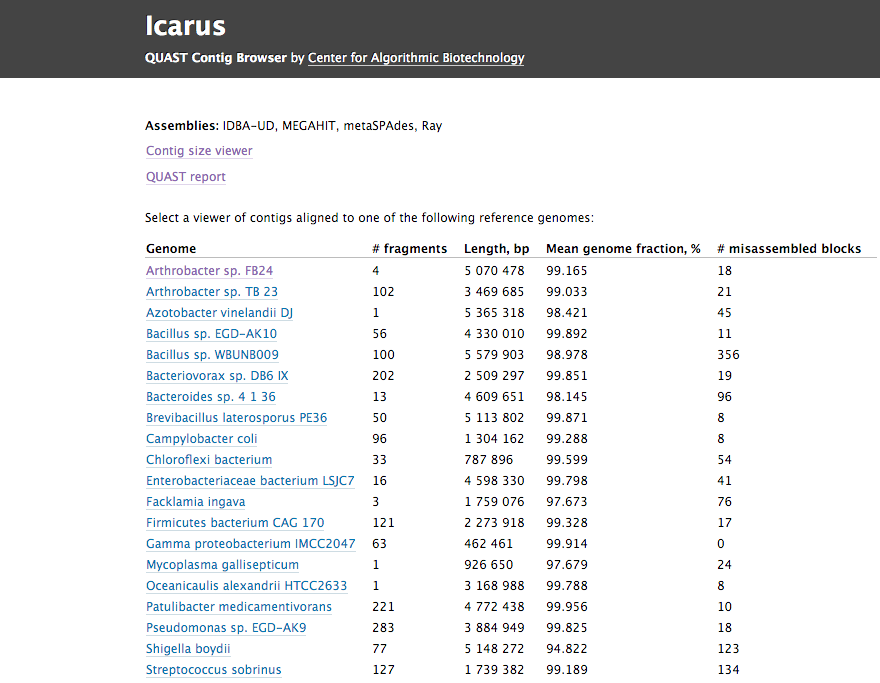
Icarus is a novel genome visualizer for accurate assessment and analysis of genomic draft assemblies, which is based on QUAST genome quality assessment tool. Icarus consists of two major contig viewers:
- Contig alignment viewer – draws contigs according to their mapping to the reference genome, visualizes misassemblies and similarities between assemblies. The viewer can visualize genes, operons, and reads coverage distribution along the genome, if any of those were provided to QUAST.
- Contig size viewer – draws contigs ordered from longest to shortest, highlights N50, N75 (NG50, NG75) and long contigs larger than a user-specified threshold.
You can try Icarus online via WebQUAST -- upload your assemblies, get regular QUAST report and press the Icarus: contig browser link.
Icarus is distributed as a command-line tool within QUAST package since version 4.0.
Icarus specific section in the Manual: output description
Citation:
Alla Mikheenko, Gleb Valin, Andrey Prjibelski, Vladislav Saveliev, Alexey Gurevich,
Icarus: visualizer for de novo assembly evaluation,
Bioinformatics (2016) 32 (21): 3321-3323. doi: 10.1093/bioinformatics/btw379
First published online: July 4, 2016
Icarus: visualizer for de novo assembly evaluation,
Bioinformatics (2016) 32 (21): 3321-3323. doi: 10.1093/bioinformatics/btw379
First published online: July 4, 2016
Examples of Icarus output on 3 datasets generated with QUAST v4.2
Please help us make QUAST better by sending your comments, bug reports, and suggestions
to alexey.gurevich@helmholtz-hips.de or
posting them on our GitHub issue tracker
(please check that an issue similar to yours is not solved or explained already!).